emmeans
Stephen Skalicky
2024-12-11
This notebook expands upon previous lessons by thinking about what
the emmeans package is actually doing when we obtain
information about a fit model. The goal is to practice interpreting main
effects and interactions, and to better understand what a marginal /
estimated mean actually…means.
library(tidyverse)
library(palmerpenguins)
library(emmeans)
data(penguins)
penguins <- drop_na(penguins)Let’s further explore how the package emmeans words to
unpack regression model effects. In doing so, we will remind ourselves
about how to interpret regression effects and interactions.
# z-score flipper length
penguins$flipper_length_mm_z <- (penguins$flipper_length_mm - mean(penguins$flipper_length_mm)) /
sd(penguins$flipper_length_mm)
summary(penguins$flipper_length_mm_z)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -2.0667 -0.7825 -0.2830 0.0000 0.8585 2.1428Let’s fit a model in with a categorical interaction between sex and species, as well as the main effect of flipper length. Do you remember how to interpret this?
##
## Call:
## lm(formula = body_mass_g ~ sex * species + flipper_length_mm_z,
## data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -832.63 -195.61 -1.39 187.86 826.25
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3638.69 50.01 72.764 < 2e-16 ***
## sexmale 580.08 49.30 11.767 < 2e-16 ***
## speciesChinstrap 77.64 60.68 1.280 0.202
## speciesGentoo 800.55 86.33 9.273 < 2e-16 ***
## flipper_length_mm_z 287.13 39.38 7.291 2.34e-12 ***
## sexmale:speciesChinstrap -335.82 84.96 -3.953 9.48e-05 ***
## sexmale:speciesGentoo 44.03 71.97 0.612 0.541
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 287.3 on 326 degrees of freedom
## Multiple R-squared: 0.875, Adjusted R-squared: 0.8727
## F-statistic: 380.2 on 6 and 326 DF, p-value: < 2.2e-16The intercept reflects the baseline level of all categorical variables and all numerical variables held at 0. So in this model, the baseline of 3638 grams is the predicted body weight of female Adelie penguins.
sexmaleis the difference between the levelmalein the variablesexand the baseline:female. Because of the interaction, this is the difference between body mass for male and female Adelie penguins. Male Adelie penguins are predicted to weight 580 grams more than female Adelie penguins.speciesChinstrapis the difference between the levelChinstrapin the variablespeciesand the baseline:Adelie. Because of the interaction, this is the difference between body mass for female Chinstrap and female Adelie penguins - Female Chinstrap penguins are predicted to weight 77 grams more than Female Adelie penguins.speciesGentoois the difference between the levelGentooin the variablespeciesand the baseline:Adelie. Because of the interaction, this is the difference between body mass for female Gentoo and female Adelie penguins - Female Gentoo pensuins are predicted to weight 800 grans more than the Female Adelie penguins.flipper_length_mm_zis the predicted effect of flipper length on body mass for the baseline (Female Adelia penguins). However, this is a main effect, meaning it will have the same effect across all levels of sex/species (see below.)
The interaction terms
sexmale:speciesChinstrapis the adjustment to predicted body mass difference between male and female Chinstrap penguins - the coefficient of -355 is subtracted from thesexmaleeffect to get the predicted difference between male/female Chinstrap penguins.sexmale:speciesGentoois the adjustment to predicted body mass differences between male and female Gentoo penguins - the coefficient of 44 is added to thesexmaleeffect to get the predicted difference between male/female Gentoo penguins.
What can we not see?
- The difference between male Adelie and male Gentoo penguins
- The difference between male Adelie and male Chinstrap penguins
- The difference between female Gentoo and female Chinstrap penguins
- The difference between the male/female difference for any pairwise comparison
confirming the average effect of flipper length
Make Gentoo the baseline, and fit a new model.
## Gentoo Adelie Chinstrap
## 119 146 68##
## Call:
## lm(formula = body_mass_g ~ sex * species + flipper_length_mm_z,
## data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -832.63 -195.61 -1.39 187.86 826.25
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 4439.24 50.11 88.583 < 2e-16 ***
## sexmale 624.12 58.25 10.715 < 2e-16 ***
## speciesAdelie -800.55 86.33 -9.273 < 2e-16 ***
## speciesChinstrap -722.91 85.58 -8.447 9.99e-16 ***
## flipper_length_mm_z 287.13 39.38 7.291 2.34e-12 ***
## sexmale:speciesAdelie -44.03 71.97 -0.612 0.541
## sexmale:speciesChinstrap -379.86 87.39 -4.347 1.85e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 287.3 on 326 degrees of freedom
## Multiple R-squared: 0.875, Adjusted R-squared: 0.8727
## F-statistic: 380.2 on 6 and 326 DF, p-value: < 2.2e-16Swap it back to Adelie
## Adelie Gentoo Chinstrap
## 146 119 68the emmeans package
The problems with interpreting the regression comparisons become even more complex once we add even more interactions.
The emmeans package lets us obtain marginal and
conditional effects of all predictors in our model.
Some terminological points:
- marginal effect: the effect of a \(x\) on \(y\) (i.e., the slope)
- the slope of a continuous \(x\)
- the conditional effect of levels of a categorical \(x\)
emmeans will first compute the average values of \(x\), then fit these averaged values using
the regression formula, to provide the effect of a predictor
when it is at its average - the estimated marginal
mean. - emmean: estimate marginal mean: the marginal
effect of \(x\) on \(y\) at the mean of \(x\) - emtrend: average slope
of continuous \(x\) on \(y\)
basic emmeans usage
The simplest way to use emmeans is to ask for predicted
values of \(y\) for a particular
variable. The syntax is emmeans(model, ~ effects).
To get the emmean of body_mass_g at
different levels of sex, we type:
## NOTE: Results may be misleading due to involvement in interactions## sex emmean SE df lower.CL upper.CL
## female 3931 25.6 326 3881 3982
## male 4414 25.5 326 4364 4464
##
## Results are averaged over the levels of: species
## Confidence level used: 0.95And the same for species…:
## NOTE: Results may be misleading due to involvement in interactions## species emmean SE df lower.CL upper.CL
## Adelie 3929 38.7 326 3853 4005
## Chinstrap 3838 37.7 326 3764 3913
## Gentoo 4751 52.5 326 4648 4855
##
## Results are averaged over the levels of: sex
## Confidence level used: 0.95And for flipper length:
why do we only get one emmean here?
Because this is the effect of flipper length on body mass when flipper length is at its mean
## flipper_length_mm_z emmean SE df lower.CL upper.CL
## 3.5e-16 4173 16.6 326 4140 4205
##
## Results are averaged over the levels of: sex, species
## Confidence level used: 0.95Of course, our model has an interaction, as the warnings have been
trying to tell us. We can model these using similar syntax as the
lm call. Here we get the predicted body mass for each level
of our interaction:
## sex species emmean SE df lower.CL upper.CL
## female Adelie 3639 50.0 326 3540 3737
## male Adelie 4219 41.3 326 4137 4300
## female Chinstrap 3716 55.7 326 3607 3826
## male Chinstrap 3961 49.4 326 3863 4058
## female Gentoo 4439 50.1 326 4341 4538
## male Gentoo 5063 68.5 326 4929 5198
##
## Confidence level used: 0.95plotting the emmeans
simply convert an emmeans call into a data frame, and it becomes easy to plot:
plot1 <- emmeans(m1, ~ sex*species)%>% as.data.frame()
ggplot(plot1, aes(y = emmean, x = species, fill = sex, color = sex)) +
geom_point() +
geom_errorbar(aes(ymin = lower.CL, ymax = upper.CL), width = .2)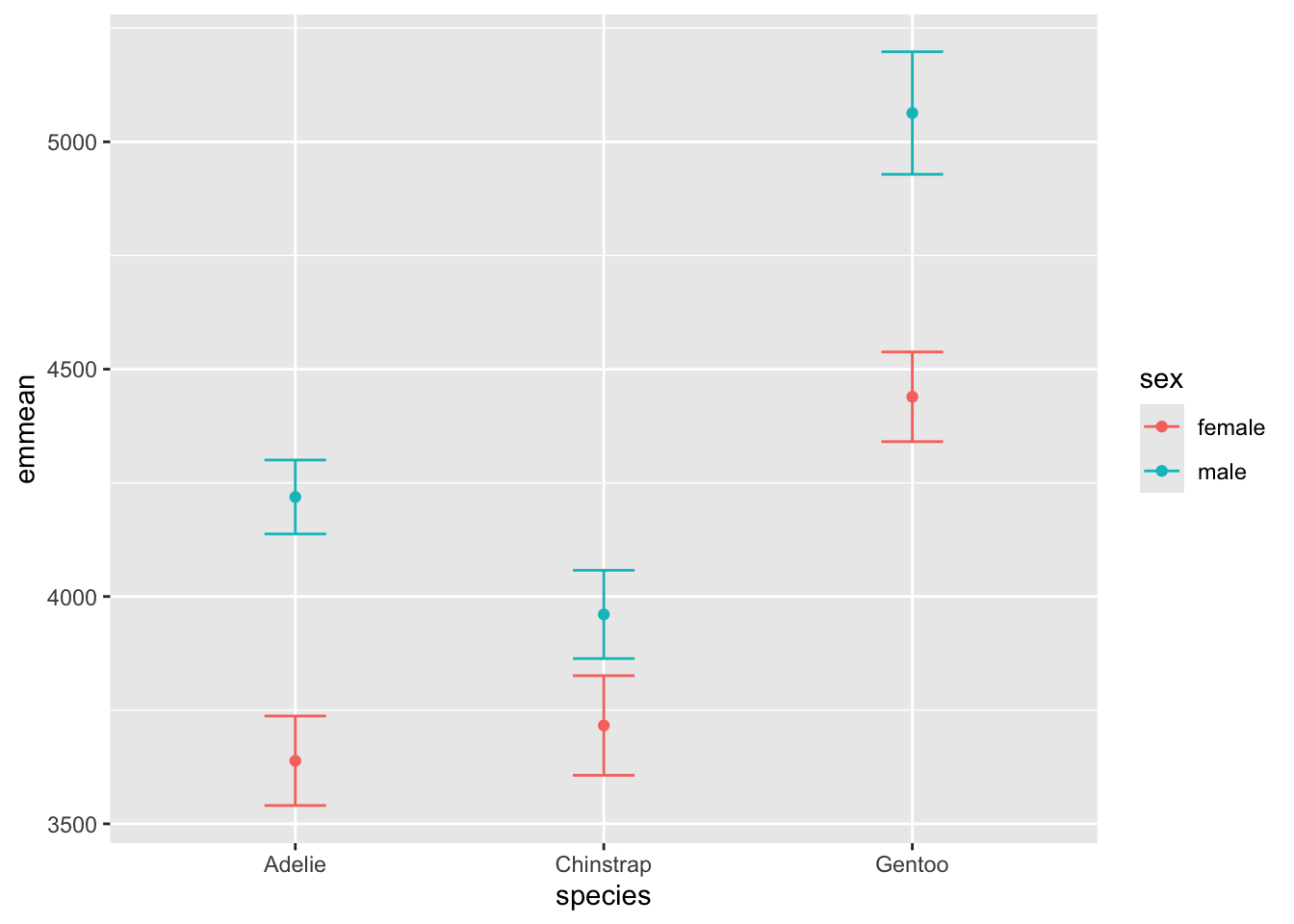
remember those pairwise comparisons?
We can think about comparisons as both within subjects
and between subjects.
The within subjects comparison will ask whether the difference in body mass between male/female penguins is “significant” for each species. The between subjects comparison could ask whether male or female penguins differ from other male or female penguins across different species
To do so, we can use the pairwise argument in
emmeans to ask for a contrast:
Pairwise contrasts of species…
## NOTE: Results may be misleading due to involvement in interactions## $emmeans
## species emmean SE df lower.CL upper.CL
## Adelie 3929 38.7 326 3853 4005
## Chinstrap 3838 37.7 326 3764 3913
## Gentoo 4751 52.5 326 4648 4855
##
## Results are averaged over the levels of: sex
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio p.value
## Adelie - Chinstrap 90.3 45.1 326 2.000 0.1139
## Adelie - Gentoo -822.6 83.8 326 -9.815 <.0001
## Chinstrap - Gentoo -912.8 74.1 326 -12.320 <.0001
##
## Results are averaged over the levels of: sex
## P value adjustment: tukey method for comparing a family of 3 estimatesPairwise contrasts of sex…
## NOTE: Results may be misleading due to involvement in interactions## $emmeans
## sex emmean SE df lower.CL upper.CL
## female 3931 25.6 326 3881 3982
## male 4414 25.5 326 4364 4464
##
## Results are averaged over the levels of: species
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio p.value
## female - male -483 38.9 326 -12.426 <.0001
##
## Results are averaged over the levels of: speciesALL the pairwise!!
Ask yourself - how many of these comparisons do we actually want?
## $emmeans
## sex species emmean SE df lower.CL upper.CL
## female Adelie 3639 50.0 326 3540 3737
## male Adelie 4219 41.3 326 4137 4300
## female Chinstrap 3716 55.7 326 3607 3826
## male Chinstrap 3961 49.4 326 3863 4058
## female Gentoo 4439 50.1 326 4341 4538
## male Gentoo 5063 68.5 326 4929 5198
##
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio p.value
## female Adelie - male Adelie -580.1 49.3 326 -11.767 <.0001
## female Adelie - female Chinstrap -77.6 60.7 326 -1.280 0.7961
## female Adelie - male Chinstrap -321.9 68.7 326 -4.686 0.0001
## female Adelie - female Gentoo -800.5 86.3 326 -9.273 <.0001
## female Adelie - male Gentoo -1424.7 107.1 326 -13.300 <.0001
## male Adelie - female Chinstrap 502.4 59.7 326 8.418 <.0001
## male Adelie - male Chinstrap 258.2 63.3 326 4.081 0.0008
## male Adelie - female Gentoo -220.5 76.2 326 -2.893 0.0464
## male Adelie - male Gentoo -844.6 95.8 326 -8.814 <.0001
## female Chinstrap - male Chinstrap -244.3 73.4 326 -3.329 0.0124
## female Chinstrap - female Gentoo -722.9 85.6 326 -8.447 <.0001
## female Chinstrap - male Gentoo -1347.0 103.9 326 -12.965 <.0001
## male Chinstrap - female Gentoo -478.6 71.7 326 -6.674 <.0001
## male Chinstrap - male Gentoo -1102.8 86.5 326 -12.755 <.0001
## female Gentoo - male Gentoo -624.1 58.2 326 -10.715 <.0001
##
## P value adjustment: tukey method for comparing a family of 6 estimatesA more sensible approach is to ask for conditional effects using the
pipe: |
Compare the between group effect of species at among females and among males (no male-female cross-species comparisons)
## sex = female:
## contrast estimate SE df t.ratio p.value
## Adelie - Chinstrap -77.6 60.7 326 -1.280 0.4077
## Adelie - Gentoo -800.5 86.3 326 -9.273 <.0001
## Chinstrap - Gentoo -722.9 85.6 326 -8.447 <.0001
##
## sex = male:
## contrast estimate SE df t.ratio p.value
## Adelie - Chinstrap 258.2 63.3 326 4.081 0.0002
## Adelie - Gentoo -844.6 95.8 326 -8.814 <.0001
## Chinstrap - Gentoo -1102.8 86.5 326 -12.755 <.0001
##
## P value adjustment: tukey method for comparing a family of 3 estimatesCompare the within group effect of sex in each species. Is it really very interesting to us that all males > females?
And, which of these effects do we see in the model output?
## species = Adelie:
## contrast estimate SE df t.ratio p.value
## female - male -580 49.3 326 -11.767 <.0001
##
## species = Chinstrap:
## contrast estimate SE df t.ratio p.value
## female - male -244 73.4 326 -3.329 0.0010
##
## species = Gentoo:
## contrast estimate SE df t.ratio p.value
## female - male -624 58.2 326 -10.715 <.0001##
## Call:
## lm(formula = body_mass_g ~ sex * species + flipper_length_mm_z,
## data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -832.63 -195.61 -1.39 187.86 826.25
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3638.69 50.01 72.764 < 2e-16 ***
## sexmale 580.08 49.30 11.767 < 2e-16 ***
## speciesChinstrap 77.64 60.68 1.280 0.202
## speciesGentoo 800.55 86.33 9.273 < 2e-16 ***
## flipper_length_mm_z 287.13 39.38 7.291 2.34e-12 ***
## sexmale:speciesChinstrap -335.82 84.96 -3.953 9.48e-05 ***
## sexmale:speciesGentoo 44.03 71.97 0.612 0.541
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 287.3 on 326 degrees of freedom
## Multiple R-squared: 0.875, Adjusted R-squared: 0.8727
## F-statistic: 380.2 on 6 and 326 DF, p-value: < 2.2e-16and compare the size of the interaction
## sex_pairwise species_pairwise estimate SE df t.ratio p.value
## female - male Adelie - Chinstrap -336 85.0 326 -3.953 0.0001
## female - male Adelie - Gentoo 44 72.0 326 0.612 0.5410
## female - male Chinstrap - Gentoo 380 87.4 326 4.347 <.0001Again, which of these can we see in the summary output?
##
## Call:
## lm(formula = body_mass_g ~ sex * species + flipper_length_mm_z,
## data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -832.63 -195.61 -1.39 187.86 826.25
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3638.69 50.01 72.764 < 2e-16 ***
## sexmale 580.08 49.30 11.767 < 2e-16 ***
## speciesChinstrap 77.64 60.68 1.280 0.202
## speciesGentoo 800.55 86.33 9.273 < 2e-16 ***
## flipper_length_mm_z 287.13 39.38 7.291 2.34e-12 ***
## sexmale:speciesChinstrap -335.82 84.96 -3.953 9.48e-05 ***
## sexmale:speciesGentoo 44.03 71.97 0.612 0.541
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 287.3 on 326 degrees of freedom
## Multiple R-squared: 0.875, Adjusted R-squared: 0.8727
## F-statistic: 380.2 on 6 and 326 DF, p-value: < 2.2e-16finally….what about flipper length?
We have the predicted body mass of penguins when flipper length is held at its mean:
## flipper_length_mm_z emmean SE df lower.CL upper.CL
## 3.5e-16 4173 16.6 326 4140 4205
##
## Results are averaged over the levels of: sex, species
## Confidence level used: 0.95To obtain the average effect of flipper length, or
more specifically, the effect of flipper length when it is held at its
mean, we use emtrends() for continuous trends (slopes)
## flipper_length_mm_z flipper_length_mm_z.trend SE df lower.CL upper.CL
## 3.5e-16 287 39.4 326 210 365
##
## Results are averaged over the levels of: sex, species
## Confidence level used: 0.95This is, of course, exactly what the regression model output will show us:
##
## Call:
## lm(formula = body_mass_g ~ sex * species + flipper_length_mm_z,
## data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -832.63 -195.61 -1.39 187.86 826.25
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 3638.69 50.01 72.764 < 2e-16 ***
## sexmale 580.08 49.30 11.767 < 2e-16 ***
## speciesChinstrap 77.64 60.68 1.280 0.202
## speciesGentoo 800.55 86.33 9.273 < 2e-16 ***
## flipper_length_mm_z 287.13 39.38 7.291 2.34e-12 ***
## sexmale:speciesChinstrap -335.82 84.96 -3.953 9.48e-05 ***
## sexmale:speciesGentoo 44.03 71.97 0.612 0.541
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 287.3 on 326 degrees of freedom
## Multiple R-squared: 0.875, Adjusted R-squared: 0.8727
## F-statistic: 380.2 on 6 and 326 DF, p-value: < 2.2e-16We can plug in a range of different values for
flipper_length_mm_z to emtrend() to see how it
moves across values in our data.
flipper_range = seq(from = min(penguins$flipper_length_mm_z), to = max(penguins$flipper_length_mm_z), by = .1)
emtrends(m1, ~ flipper_length_mm_z, var = 'flipper_length_mm_z', at = list(flipper_length_mm_z = flipper_range))## flipper_length_mm_z flipper_length_mm_z.trend SE df lower.CL upper.CL
## -2.0667 287 39.4 326 210 365
## -1.9667 287 39.4 326 210 365
## -1.8667 287 39.4 326 210 365
## -1.7667 287 39.4 326 210 365
## -1.6667 287 39.4 326 210 365
## -1.5667 287 39.4 326 210 365
## -1.4667 287 39.4 326 210 365
## -1.3667 287 39.4 326 210 365
## -1.2667 287 39.4 326 210 365
## -1.1667 287 39.4 326 210 365
## -1.0667 287 39.4 326 210 365
## -0.9667 287 39.4 326 210 365
## -0.8667 287 39.4 326 210 365
## -0.7667 287 39.4 326 210 365
## -0.6667 287 39.4 326 210 365
## -0.5667 287 39.4 326 210 365
## -0.4667 287 39.4 326 210 365
## -0.3667 287 39.4 326 210 365
## -0.2667 287 39.4 326 210 365
## -0.1667 287 39.4 326 210 365
## -0.0667 287 39.4 326 210 365
## 0.0333 287 39.4 326 210 365
## 0.1333 287 39.4 326 210 365
## 0.2333 287 39.4 326 210 365
## 0.3333 287 39.4 326 210 365
## 0.4333 287 39.4 326 210 365
## 0.5333 287 39.4 326 210 365
## 0.6333 287 39.4 326 210 365
## 0.7333 287 39.4 326 210 365
## 0.8333 287 39.4 326 210 365
## 0.9333 287 39.4 326 210 365
## 1.0333 287 39.4 326 210 365
## 1.1333 287 39.4 326 210 365
## 1.2333 287 39.4 326 210 365
## 1.3333 287 39.4 326 210 365
## 1.4333 287 39.4 326 210 365
## 1.5333 287 39.4 326 210 365
## 1.6333 287 39.4 326 210 365
## 1.7333 287 39.4 326 210 365
## 1.8333 287 39.4 326 210 365
## 1.9333 287 39.4 326 210 365
## 2.0333 287 39.4 326 210 365
## 2.1333 287 39.4 326 210 365
##
## Results are averaged over the levels of: sex, species
## Confidence level used: 0.95That gives us a lot of values…better to plot it? We can use the
emmip function to make a plot grid.
# pretty!
flipper_plot <- as.data.frame(emmip(m1, ~ flipper_length_mm_z, var = 'flipper_length_mm_z', at = list(flipper_length_mm_z = flipper_range), CIs = TRUE, plotit = F))
ggplot(flipper_plot, aes(x = flipper_length_mm_z, y = yvar)) +
geom_ribbon(aes(ymin = LCL, ymax = UCL), alpha = .2) +
geom_line() +
labs(y = 'Body Mass (g)', x = 'Flipper Length (z-scored)')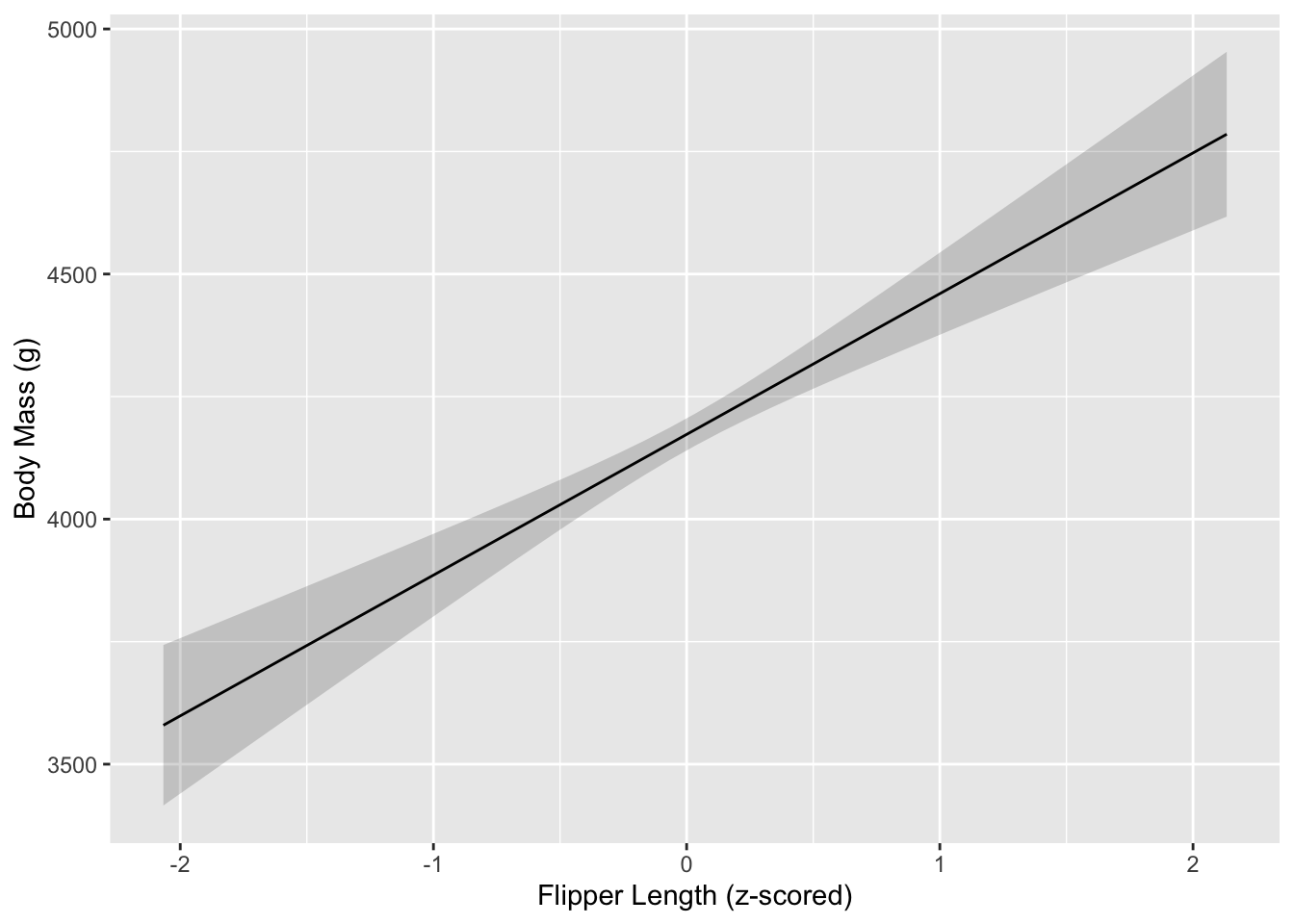
But we can also use emmeans to get predicted body mass at different levels of flipper length…
## flipper_length_mm_z emmean SE df lower.CL upper.CL
## -2.0667 3579 83.2 326 3416 3743
## -1.9667 3608 79.3 326 3452 3764
## -1.8667 3637 75.5 326 3488 3785
## -1.7667 3666 71.7 326 3525 3807
## -1.6667 3694 67.8 326 3561 3828
## -1.5667 3723 64.0 326 3597 3849
## -1.4667 3752 60.2 326 3633 3870
## -1.3667 3780 56.5 326 3669 3891
## -1.2667 3809 52.7 326 3705 3913
## -1.1667 3838 49.0 326 3741 3934
## -1.0667 3867 45.3 326 3777 3956
## -0.9667 3895 41.7 326 3813 3977
## -0.8667 3924 38.1 326 3849 3999
## -0.7667 3953 34.6 326 3885 4021
## -0.6667 3981 31.2 326 3920 4043
## -0.5667 4010 27.9 326 3955 4065
## -0.4667 4039 24.9 326 3990 4088
## -0.3667 4068 22.1 326 4024 4111
## -0.2667 4096 19.7 326 4057 4135
## -0.1667 4125 17.9 326 4090 4160
## -0.0667 4154 16.8 326 4121 4187
## 0.0333 4182 16.6 326 4150 4215
## 0.1333 4211 17.3 326 4177 4245
## 0.2333 4240 18.9 326 4203 4277
## 0.3333 4269 21.1 326 4227 4310
## 0.4333 4297 23.7 326 4251 4344
## 0.5333 4326 26.6 326 4274 4378
## 0.6333 4355 29.8 326 4296 4413
## 0.7333 4383 33.2 326 4318 4449
## 0.8333 4412 36.6 326 4340 4484
## 0.9333 4441 40.2 326 4362 4520
## 1.0333 4470 43.8 326 4383 4556
## 1.1333 4498 47.5 326 4405 4592
## 1.2333 4527 51.2 326 4426 4628
## 1.3333 4556 54.9 326 4448 4664
## 1.4333 4584 58.7 326 4469 4700
## 1.5333 4613 62.5 326 4490 4736
## 1.6333 4642 66.3 326 4511 4772
## 1.7333 4670 70.1 326 4533 4808
## 1.8333 4699 73.9 326 4554 4845
## 1.9333 4728 77.8 326 4575 4881
## 2.0333 4757 81.6 326 4596 4917
## 2.1333 4785 85.5 326 4617 4954
##
## Results are averaged over the levels of: sex, species
## Confidence level used: 0.95Plotting this shows us that…
# it's the exact same plot!!!!
p3 <- as.data.frame(emmeans(m1, ~ flipper_length_mm_z, at = list(flipper_length_mm_z = flipper_range)))
ggplot(p3, aes(y = emmean, x = flipper_length_mm_z)) +
geom_ribbon(aes(ymin = lower.CL, ymax = upper.CL), alpha = .2) +
geom_line()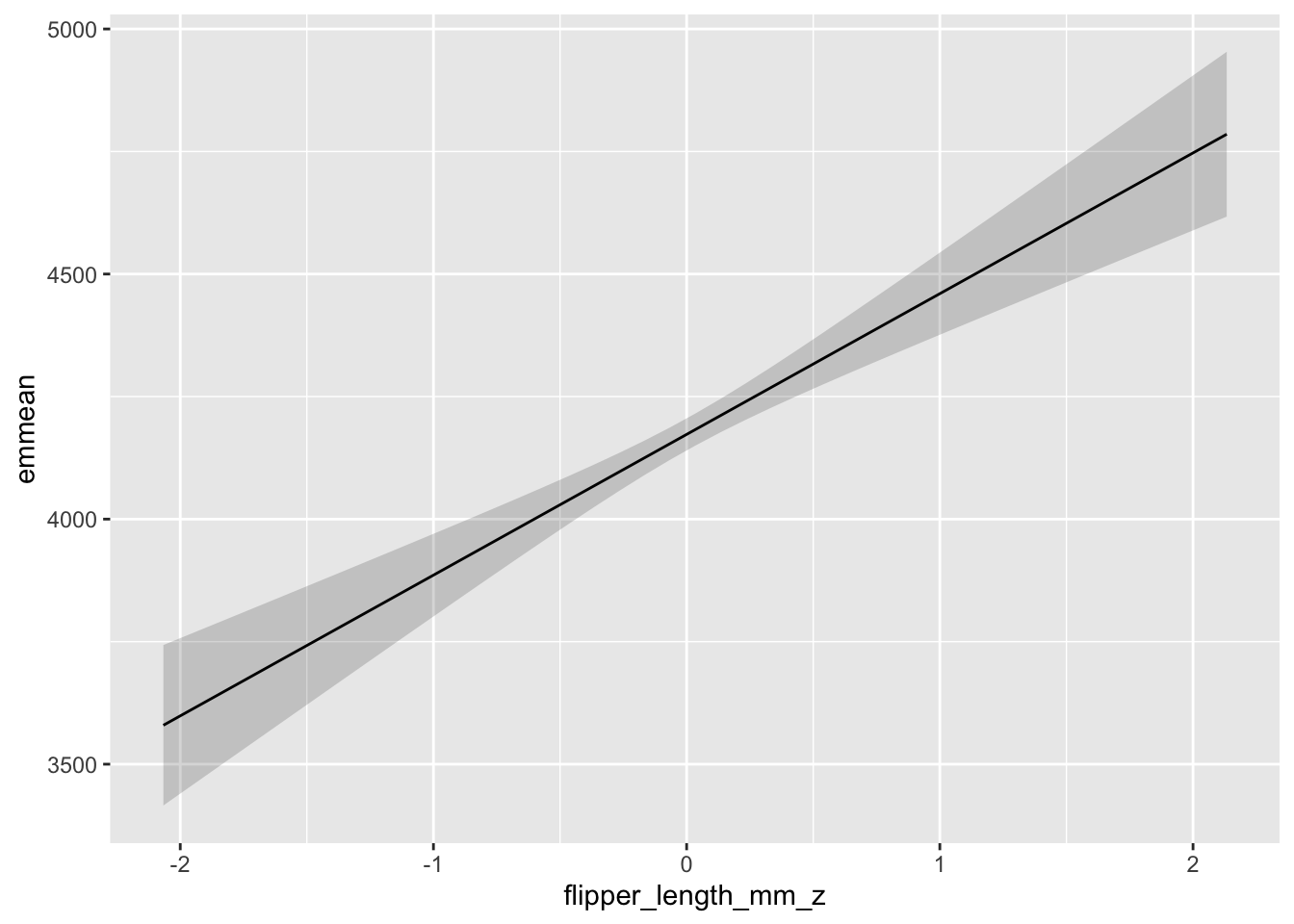
Actually, the average effect from emmeans is just one
way of doing these things - it provides the answer to the “average case”
in our data. This is slightly different than the actual average effect
in our population…but that requires a different approach. Maybe for next
time!